People are often curious about how their bodies work. So, it is no surprise that single-cell RNA sequencing (scRNA-seq) — which has the power to map all the cell types in the human body1–3 — has drawn great interest from scientists and funding agencies alike. But a major limitation of scRNA-seq is that it cannot provide information about where in the original tissue each cell was located. Writing in Cell, Asp et al.4 demonstrate a way of overcoming this hurdle by combining scRNA-seq with other sequencing methods that retain location information. They use this approach to create a spatially defined cell atlas of the developing human heart.
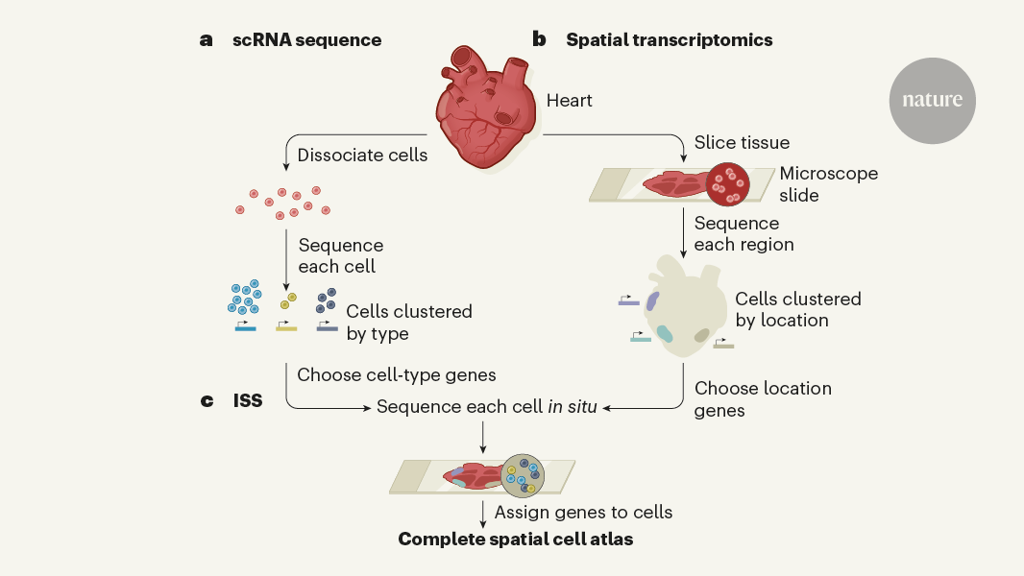
ScRNA-seq involves dissociation of a tissue into hundreds or thousands of individual cells, each of which is analysed to determine its gene-expression profile. This profile indicates the proteins and pathways that are active in that cell — information that computational methods can then use to sort thousands of cells into different types or states at once. However, the tissue-dissociation protocol breaks the link between single cells and their original positions in the tissue. Without this information, interpretation of the data is incomplete. Asp et al. set out to bridge this gap by combining scRNA-seq with two approaches that produce spatially defined gene-expression maps, although at a lower resolution than scRNA-seq.
The first is a technique called spatial transcriptomics5. Thin slices of tissue are placed on a specially prepared microscope slide that has been dotted with circular ‘patches’. Each patch contains many copies of a nucleic-acid probe that binds to messenger RNA in the tissue sample and carries a sequence called a barcode. Each patch has a different barcode, so that a specific label is attached to the mRNA in the area of tissue sitting on top of that patch. When the mRNA from each region is sequenced, the barcode acts as a record of the cells’ original locations in the tissue. A limitation of this method is that single-cell analysis is compromised, because gene sequences are pooled from approximately 30 cells in each patch. Nonetheless, a non-biased description of gene-expression profiles at discrete locations is obtained.
The second method is in situ sequencing (ISS), in which the expression of preselected genes is probed directly on a tissue slice on a microscope slide6,7. DNA probes are designed to bind to mRNA transcribed from genes of interest, with each probe carrying a unique barcode. When these probes are introduced into a tissue, they bind to their target mRNAs in the tissue’s cells and — after further processing steps — a fluorescent-imaging-based sequencing method is used to detect the barcodes while they remain in place in the tissue. This technique can provide information about the expression of 50–100 genes in individual cells at high spatial resolution. But the genes must be preselected, which requires some knowledge about which genes will be informative.
Asp and colleagues ultimately used ISS to achieve cell-level mapping of heart development. But to determine which genes they should select for ISS, they needed to perform scRNA-seq and spatial transcriptomics (Fig. 1). The authors’ scRNA-seq analysis revealed genes correlating with cell type — genes expressed only in smooth-muscle cells, for instance. And their spatial transcriptomics analysis revealed groups of genes that broadly correlated with certain locations in the heart — genes specifically expressed in cells in a region called the outflow tract, for example.
The researchers then created a panel of 69 ISS probes — some corresponding to the location markers identified by spatial transcriptomics, some to the cell-type markers identified by scRNA-seq, and some to genes previously reported to be important for heart development. They combined the data produced by this ISS screen with their scRNA-seq data using a recently developed algorithm8 that assigns each RNA molecule detected by ISS to a specific cell nucleus, and each nucleus to a specific cell type. The final result was the first spatiotemporal cell atlas of the embryonic human heart between 6.5 and 7 weeks of development. Anybody can explore the atlas using a searchable online tool (see go.nature.com/3rj6dtf).
Beyond a technical proof-of-principle, the authors also gained insights into heart development. For example, a previous study9 described several distinct, but transcriptionally similar, clusters of fibroblast-like cells — a structural cell type that is not fully understood, but which is known to participate in a pathological tissue-scarring process called fibrosis. Asp et al. demonstrated that these fibroblast subgroups are located in distinct parts of the heart, providing a clue as to how they might function differently.
As another example, Asp and colleagues described a previously unknown human equivalent of a subpopulation of cardiac muscle cells found in mice that expresses high levels of the gene Myoz210. Finally, the researchers performed a cellular analysis of a tissue called the atrioventricular mesenchyme, and identified the time point at which Schwann cells (neuron-associating cells that ensure proper electrical transmission) arise in this tissue. Such knowledge of human heart cells can be used to inform follow-up experiments aimed at defining those cells’ functions.
However, limitations in Asp and colleagues’ study highlight that pre-existing knowledge is still required to fully appreciate single-cell data. For instance, some of their cell populations seem to be misidentified. One population is deemed by the authors to be cells that line capillary blood vessels (coronary endothelium). But on the basis of the cells’ marker-gene expression and location, we would suggest that they are instead endocardial cells that line the heart chamber. Furthermore, the authors’ approach could not be used to distinguish between all of the subtypes or substates in particular populations, such as the coronary endothelium, which is a mixed population that lines the arterial, venous and capillary blood vessels leading to and from the heart. However, the authors’ workflow is certain to be refined as advanced techniques are developed for directly profiling the spatial gene expression of tissues11,12.
A major strength of Asp and colleagues’ study is that it provides a framework for maximizing the power of scRNA-seq. Anyone who begins experiments to understand human biology faces the challenge of limited sample availability, particularly when assessing embryonic development. The techniques presented here have the crucial advantage of providing a wealth of information from limited tissue. Asp and co-workers’ strategy is likely to benefit efforts such as the Human Cell Atlas and HuBMAP projects, which are both seeking to fully define the human body.